
We suggest to quantify the samples via fluorometry (Qubit or plate reader) for accurate pooling. Once you have verified (via agarose gel electrophoresis) that the PCR products for all samples are clean and of about the same and expected size, the samples should be pooled equimolarly. The PCR reactions should be cleaned up with Ampure XP beads (or similar) and resuspended in EB buffer. Please optimize the conditions of the first round PCR to avoid primer-dimer generation. P7-PCR index primer: 5’ CAAGCAGAAGACGGCATACGAGATGTCTCGTGGGCTCGG P5-PCR index primer: 5’ AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTC The second round PCR primers are Nextera-style index primers – i5 and i7 indicate the location of the barcode index sequences: Reverse overhang P7-tag: 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG. The first round PCR primer designs use Nextera-style tag sequences (overhang sequences) and look like this:įorward overhang P5-tag: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG. However, for HiSeq 4000 and NovaSeq sequencing you should use uniquely-dual-indexed (UDI) barcode combinations. If you are using single indices they have to be i7 (P7 adapter) indices. Since you can use dual indexes, you could order for example 5 index oligos with i5 indexes and 5 index oligos with i7 indexes and have 25 usable barcode combinations for your project. Your first round PCR amplicon products will have universal tails/tags/overhangs on both ends. We strongly recommend using plates with single-reaction aliquots of these index primers for your experiments to make sure that index primer stocks cannot become contaminated. Thus, low-cost desalted oligos can be ordered for this purpose anywhere and will work just fine. The oligos are used for standard PCR reactions. When ordering oligos please use the index sequences in the “Bases in Adapter” columns.
These indices allow for the combinatorial sequencing of up to 468 samples. The sequences for the index primers (26 i7 index 1 sequences 18 i5 index 2 sequences) are available on pages 7 and 8 here. There is no need to purchase an Illumina Nextera index kit. It is advisable to avoid any sequences that generate a Delta G smaller than -9 for any of the structures.
#ILLUMINA ADAPTER SEQUENCES PLUS#
Once you have designed the oligos as described in the Illumina protocol (forward overhang plus your sequence-specific primer as well as reverse overhang plus sequence-specific primer), we suggest checking these sequences on the IDT oligoanalyzer ( ) for secondary structures.
#ILLUMINA ADAPTER SEQUENCES FULL#
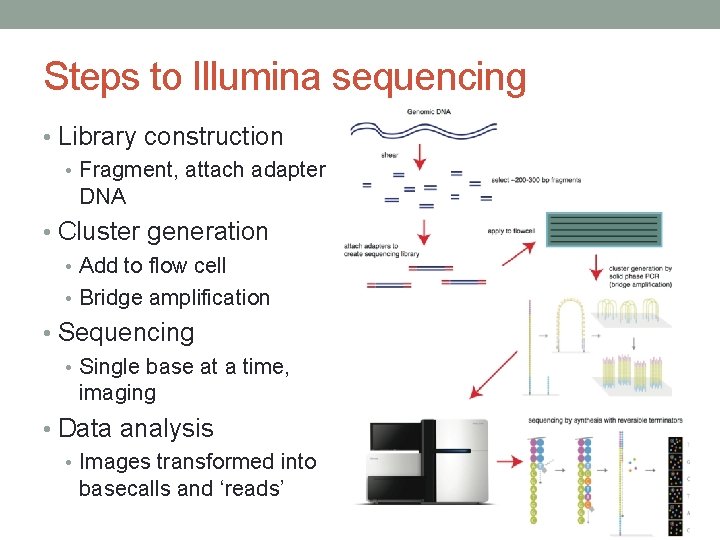
We suggest to follow a “16S amplicon” protocol that was explicitly designed by Illumina to be adaptable to other targets (please see the full protocol and pages 3 and 4 here).

This protocol has the advantage that it does not require custom sequencing primers and that the barcode-indexing oligos can be re-used for multiple different amplicons and future projects. Here we describe one of many options: A two-step PCR protocol to generate complete sequencing libraries. There are multiple valid protocols available for amplicon sequencing on Illumina systems.


 0 kommentar(er)
0 kommentar(er)
